In previous posts we talked about “Together Against PRRS”, a global integrated program developed by HIPRA for the control of PRRSv. In this entry we’ll focus on the results of the systematic monitoring program and its impact on the control of PRRS on pig farms within a Spanish large production group.
A total of 40 Spanish breeding herds enrolled in the program.
All of them were classified following the Holtkamp classification system (Holtkamp et al., 2011) following the same diagnostic monitoring protocol: monthly individual blood sampling of 30 due-to-wean piglets.
Samples were pooled and tested for PRRSv RNA by RT-PCR.
As we explained in the previous post, some PCR-positive result samples were also sequenced with the objective of describing the PRRSv genetic diversity among the study herds.
What was the trend in terms of PRRSv status?
13 samplings per farm were performed during the one-year monitoring period on the 35 PRRSv positive farms included in the study.
Farm classification trends according to PRRS status (PUS: PRRS positive unstable; PSS: PRRS positive stable) are shown in Figure 1.
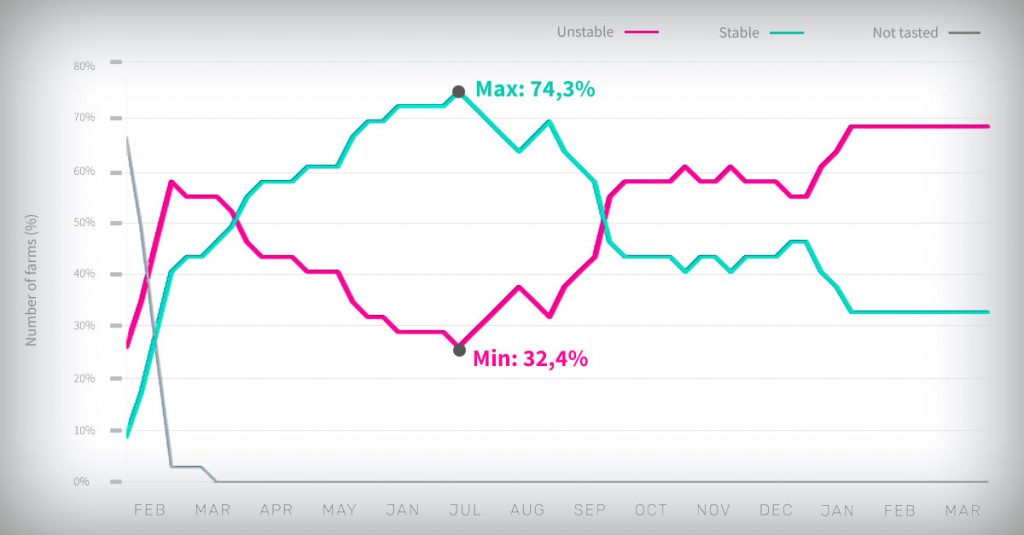
Figure 1. Trends in the percentage of unstable (PUS) and stable (PSS) farms during the monitoring period (February 2017 to March 2018).
As shown in the figure above, the percentage of PSS farms increased from the beginning of the study until mid-July 2017 when it reached the maximum (74.3%).
Afterwards, PSS farms decreased until January 2018 when they reached the minimum (32.4%).
What information did sequenced PRRS PCR-positive samples give us?
According to the sequences obtained from 24 different farms by sequence homology and phylogenetic analysis, we were able to identify 8 different PRRSv clusters encompassing 40 of the sequences but 5 sequences could not be related to any of these clusters.
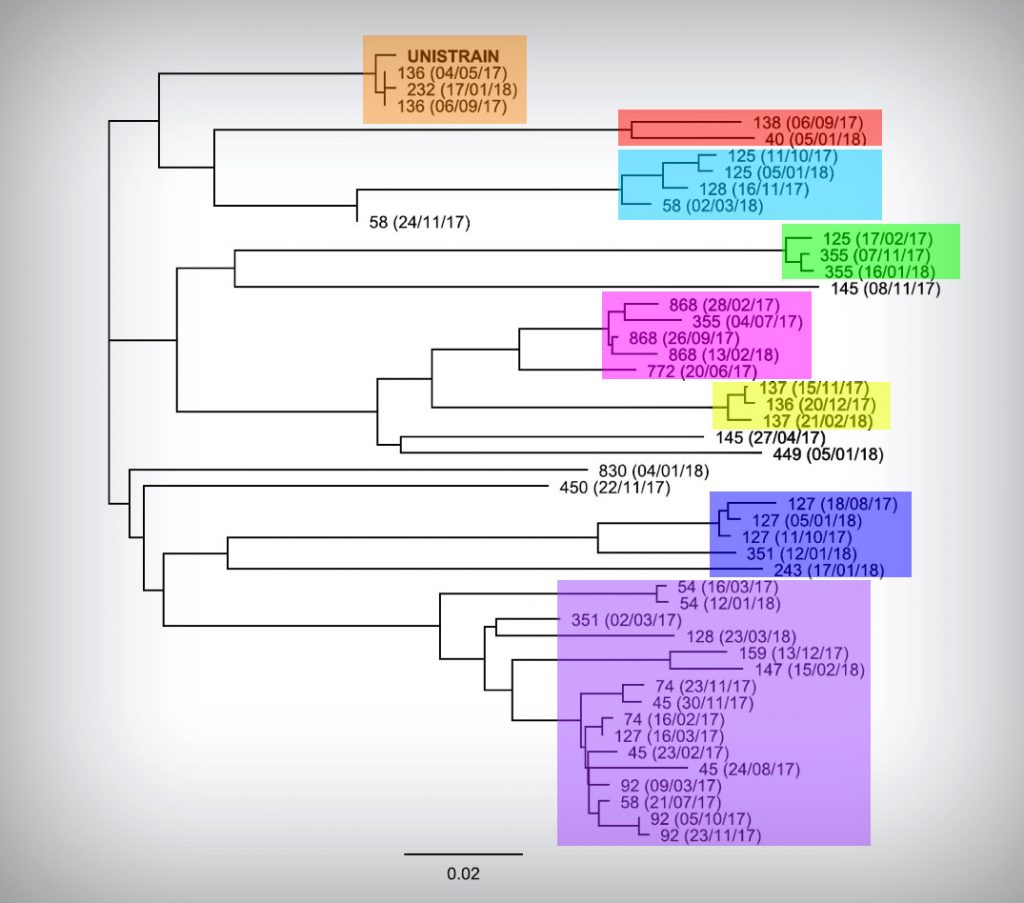
Figure 2. Phylogenetic analysis of the PRRSv strains sequenced. Sample references: farm code (sampling date). Groups of strains in the same color box indicate a strong phylogenetic relationship (% homology > 95%).
Amongst the results, we were be able to sequence just one PRRSv strain on 9 farms and more than one different PRRSv strain on 15 farms.
On the farms where we obtained more than one strain:
- We observed recirculation events of the same PRRSv field strain on 10 farms.
- Lateral introduction of a new PRRSv strain on to 8 farms.
- Both events (recirculation and introduction of new PRRSv strains) on 3 farms.
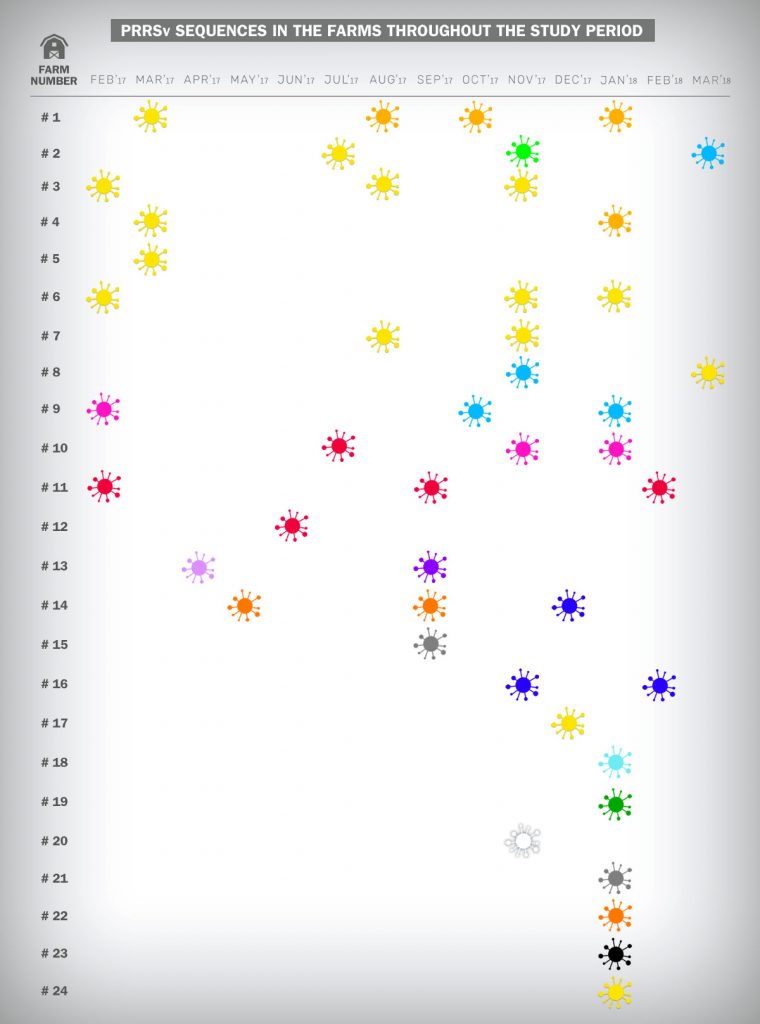
Figure 3. Epidemiologic study of the PRRS strains obtained by sequencing on the farms throughout the study period. The same color represents the same strain sequence. A different color represents a different PRRS strain obtained by sequencing.
On two farms, the nucleotide sequence of the sample matched the one for the MLV applied in the sows.
All together this indicates a highly dynamic PRRS epidemiology within the farms in the group, frequently with both PRRSv new infections and recirculation.
Most of the PRRSv strains identified through ORF5 nucleotide sequencing were included in one of the clusters described in this study (purple box in figure 2) and showed a very close relationship with others strains found on different farms at different times.
This could be explained by the geographical location of the farms within the group. They were all located in north-eastern regions of Spain (Cataluña, Aragón and Navarra) with the highest density of pig farms in Spain and an area with the most intensive pig production and slaughtering activity in the country.
These results suggest that both PRRSv recirculation and lateral infections can play a critical role for the control of PRRS on pig farms.
So, internal and external biosecurity are both key points in PRRSv control strategies.
At the same time, these findings indicate that PRRSv strain-sharing can very often be found among farms in the same production group or system, indicating a significant epidemiologic relationship between them.
Altogether, it reinforces the necessity to include PRRSv sequencing as a systematic tool for a better understanding of the epidemiologic relationships between farms of the same production systems or that are closely located.
In the next post, we will explain the impact of the routine vaccination programs applied to sows and piglets during this project in the farm´s PRRS status classification.
REFERENCES:
- Holkamp et al. Terminology for classifying swine herds by porcine reproductive and respiratory syndrome virus status. J Swine Health and Prod. 2011;19 (1): 44-56.
HIPRA © All rights reserved




