Identification of PRRS virus can be accomplished by the detection of viral proteins (antigen), by virus isolation and by the detection of nucleic acids.
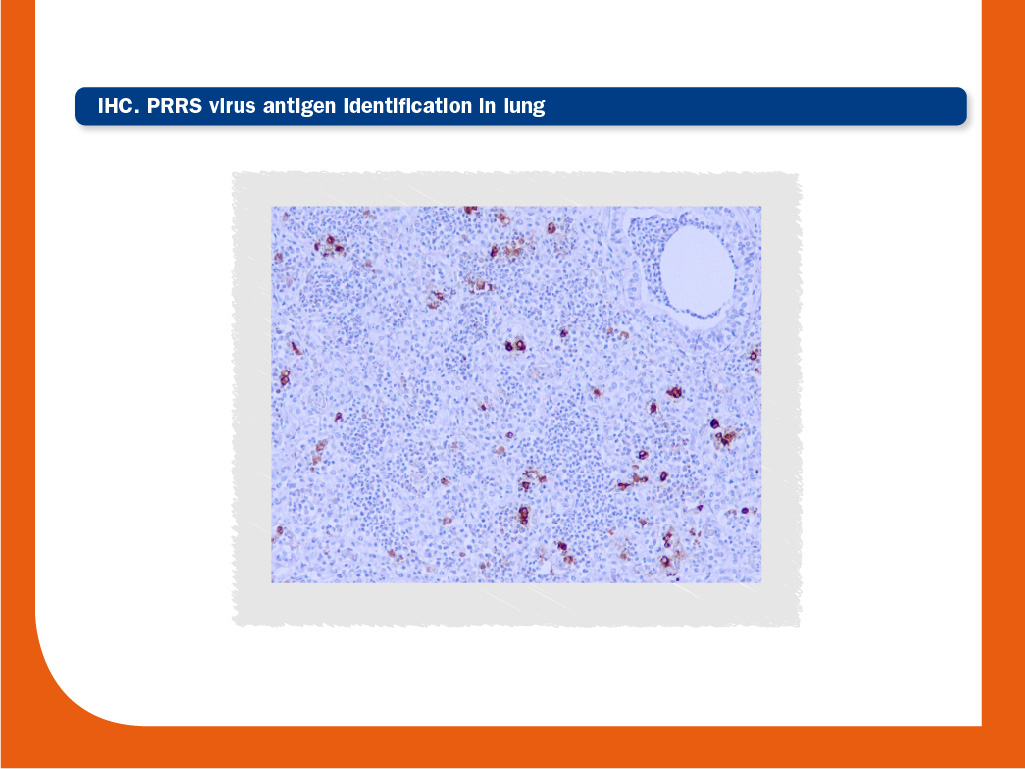
Immunohistochemistry (IHC) and in-situ hybridization (ISH)
PRRS virus antigen or nucleic acid can be detected in tissues by IHC and ISH, respectively. The relationship between viral detection and lesions can be established by a combination of IHC or ISH and histopathology.
Virus isolation and titration
Whereas PRRSV2 strains can be isolated in both porcine alveolar macrophages and sublines of the African monkey kidney cell line (CL-2621 and MARC-145), some PRRSV1 and European-like viruses can only be isolated in porcine alveolar macrophages.
Since cytopathic effects, which occur in 1-4 days, are not always clear, virus isolation usually needs to be confirmed by RT-PCR, IPMA or IFA (maximum sensitivity at 7 days). Virus titration can be done using serial dilutions of sample.
Detection of PRRS virus genome
Restriction Fragment Length Polymorphism (RFLP)
It involves ORF5 digestion by three restriction enzymes. As a result, a three-digit pattern is generated. This technique is used to differentiate isolates, however it has been demonstrated that different isolates can share similar RFLP patterns.
Therefore, sequencing is better than RFLP to differentiate isolates.
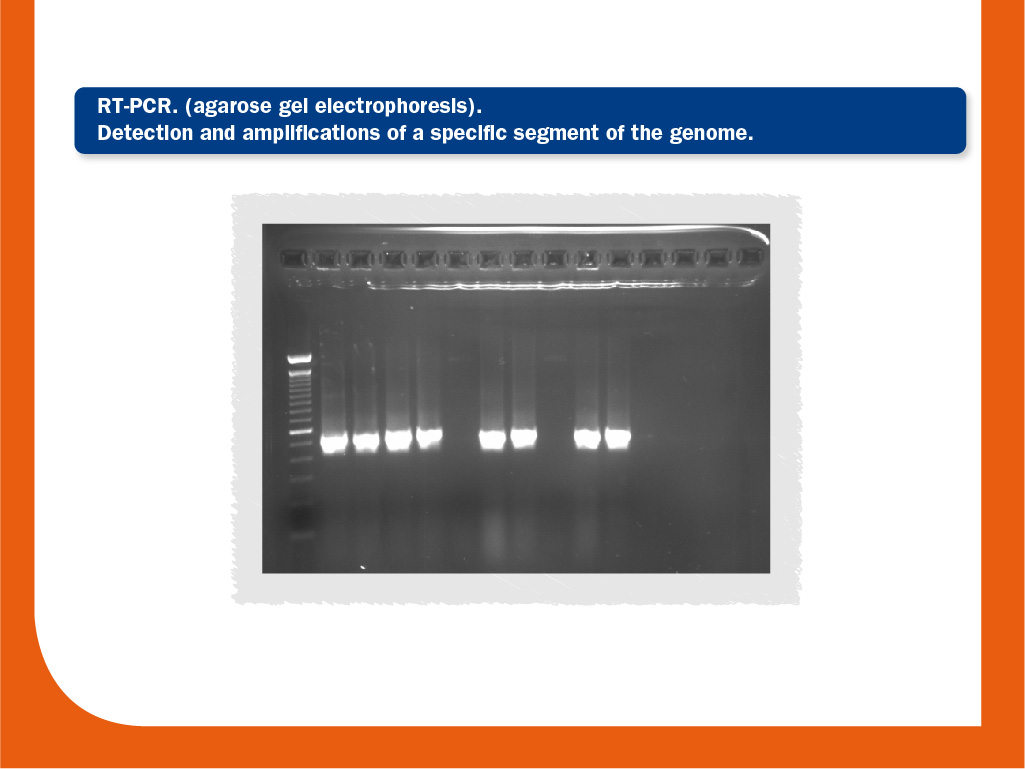
Conventional reverse transcription-polymerase chain reaction (RT-PCR)
RT-PCR, together with ELISA, is the most commonly performed diagnostic test for PRRS. It is based on the detection and the amplification of a specific segment of the genome. It is more sensitive than virus isolation, but since it detects the genome, a positive result RT-PCR does not imply the presence of viable virus.
RT-PCR is usually used to amplify ORF5 and ORF7 regions. Its sensitivity is variable since the genetic diversity of the virus is in a continuous ever-expanding mode; this phenomenon require a constant updating of the primers used in order to minimize the presence of false negative results.
It is based on the detection, the amplification and the quantification of a specific segment of the genome. It detects and measures the amplification of a targeted segment during each cycle of a PCR reaction in real-time, and not at its end, as in conventional RT-PCR.
Briefly, oligonucleotides labelled with a fluorescent reporter permits detection after hybridization of the probe with its complementary sequence.
A real time RT-PCR instrument is responsible to measure the accumulation of the fluorescent signal during amplification. A relative (semi-quantification) or absolute quantification of the targeted segment can be performed.
When an exact quantification is needed, a standard line from serial dilutions of the same strain that we intend to detect is mandatory.
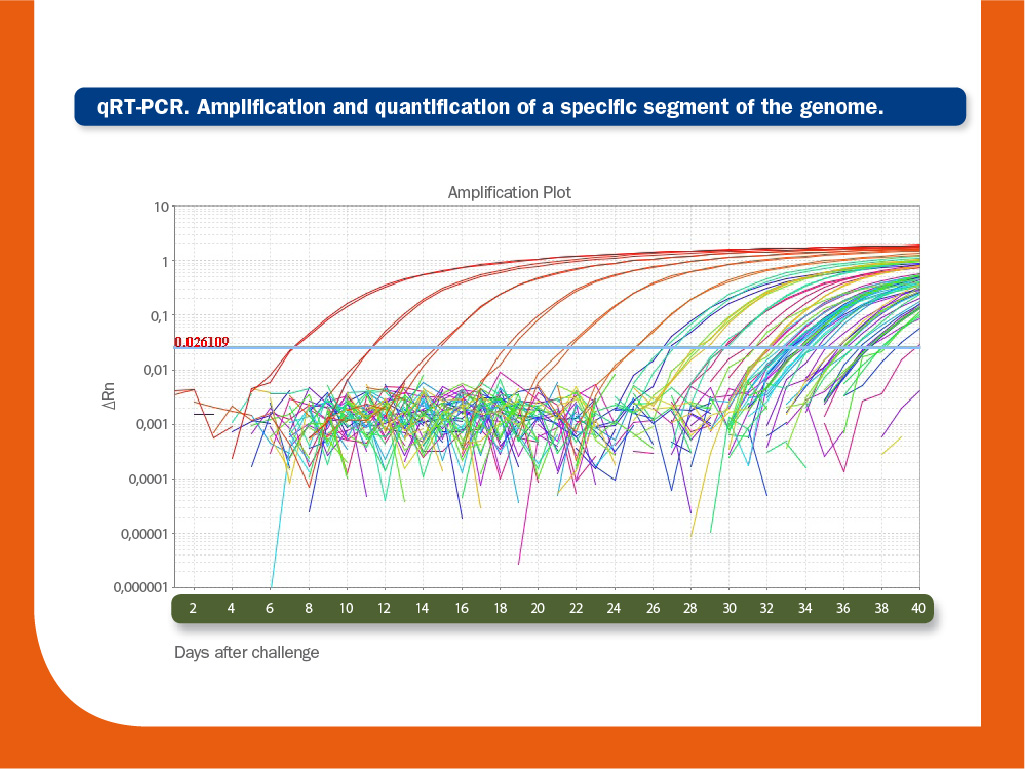
Real-time RT-PCR
RT-PCR results are usually shown as Ct values. Ct value is inverse to the amount of nucleic acid that is present in the sample.
It can be exactly (absolute quantification) or approximately (semi-quantification) correlated to the number of copies in the sample.
Therefore, lower Ct values indicate high amounts, while higher Ct values mean low amounts of the targeted nucleic acid. For PRRS virus infections, typical Ct values are around 26 cycles, with a significant variability depending on the age of the animal; i.e. Ct values around 17-18 cycles can be found in some new-born viremic piglets, whereas highest Ct values -around 26- can be found in adults.
In general, Ct values ≥ 37 cycles are considered as results with a relative low or null biological significance.
Viraemias due to vaccinations with attenuated virus usually range from 30 to 35, depending on the strain and on the age of the animal.
As occurs with conventional RT-PCR, there is a lack of uniform assay performance among labs. At present, commercial real time RT-PCR assays are available in some countries, but in some labs it is a home-made assay.
Similarly to conventional RT-PCR, a variable number of false negatives could exist; usually depending on the standardizing, improving and updating of the assay and on the expertise of the lab.
Nevertheless, sensitivity is similar or slightly higher than conventional RT-PCR, but again relies on the matching of primers and proves to the target sequences. Obviously, it can be designed to distinguish species.
© Laboratorios Hipra, S.A. 2024. All Rights Reserved.
No part of this website or any of its contents may be reproduced, copied, modified or adapted, without the prior written consent of HIPRA.
- Bautista EM, Meulenberg JJ, Choi CS, Molitor TW. Structural polypeptides of the American (VR-2332) strain of porcine reproductive and respiratory syndrome virus. Arch Virol. 1996:1357-65.
- Benfield DA, Nelson E, Collins JE, Harris L, Hennings JC, Shaw DP, Goyal SM, McCullough S, Morrison RB, Joo HS, Gorcyca D, Chladek D. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J Vet Diagn Invest. 1992. 4, 127-133.
- Benfield D, Nelson J, Rossow K, Nelson C, Steffen M, Rowland R. Diagnosis of persistent or prolonged porcine reproductive and respiratory syndrome virus infections Vet Res. 2000, 31:71.
- Brockmeier SL, Halbur PG, Thacker EL. Porcine respiratory disease complex. In KA Brogden, JM Guthmiller, eds, Polymicrobial Diseases. Washington, DC: ASM Press. 2002, 231-58.
- Calzada-Nova G, Schnitzlein W, Husmann R, Zuckermann FA. Characterization of the cytokine and maturation responses of pure populations of porcine plasmacytoid dendritic cells to porcine viruses and toll-like receptor agonists. Vet Immunol Immunopathol. 2010, 135:20-33.
- Chen WY, Schniztlein WM, Calzada-Nova G, Zuckermann FA. Genotype 2 strains of porcine reproductive and respiratory syndrome virus dysregulate alveolar macrophage cytokine production via the unfolded protein response. J Virol.2017, pii: JVI.01251-17.
- Christopher-Hennings J, Nelson EA, Hines RJ, Nelson JK, Swenson SL, Zimmerman JJ, Chase CL, Yaeger MJ, Benfield DA. Persistence of porcine reproductive and respiratory syndrome virus in serum and semen of adult boars. J Vet Diagn Invest. 1995, 7:456-64.
- Cortey M, Díaz I, Martín-Valls GE, Mateu E. Next-generation sequencing as a tool for the study of Porcine reproductive and respiratory syndrome virus (PRRSV) macro- and micro- molecular epidemiology. Vet Microbiol. 2017. doi: 10.1016/j.vetmic.2017.02.002.
- Darwich L, Díaz I, Mateu E. Certainties, doubts and hypotheses in porcine reproductive and respiratory syndrome virus immunobiology. Virus Res. 2010, 154:123-32.
- Díaz I, Darwich L, Pappaterra G, Pujols J, Mateu E. Different European-type vaccines against porcine reproductive and respiratory syndrome virus have different immunological properties and confer different protection to pigs. Virology. 2006, 351:249-59.
- Díaz I, Gimeno M, Darwich L, Navarro N, Kuzemtseva L, López S, Galindo I, Segalés J, Martín M, Pujols J, Mateu E. Characterization of homologous and heterologous adaptive immune responses in porcine reproductive and respiratory syndrome virus infection. Vet Res. 2012, 19:43:30.
- García-Nicolás O, Quereda JJ, Gómez-Laguna J, Salguero FJ, Carrasco L, Ramis G, Pallarés FJ. Cytokines transcript levels in lung and lymphoid organs during genotype 1 Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) infection. Vet Immunol Immunopathol. 2014, 160:26-40.
- Gimeno M, Darwich L, Diaz I, de la Torre E, Pujols J, Martín M, Inumaru S, Cano E, Domingo M, Montoya M, Mateu E. Cytokine profiles and phenotype regulation of antigen presenting cells by genotype-I porcine reproductive and respiratory syndrome virus isolates. Vet Res. 2011, 42:9.
- Gómez-Laguna J, Salguero FJ, Pallarés FJ, Carrasco L. Immunopathogenesis of porcine reproductive and respiratory syndrome in the respiratory tract of pigs. Vet J. 2013, 195:148-55.
- Haynes JS, Halbur PG, Sirinarumitr T, Paul PS, Meng XJ, Huffman EL. Temporal and morphologic characterization of the distribution of porcine reproductive and respiratory syndrome virus (PRRSV) by in situ hybridization in pigs infected with isolates of PRRSV that differ in virulence. Vet Pathol. 1997, 34:39-43.
- Hill H. Overview and history of Mystery Swine Disease (swine infertility/respiratory syndrome). Proceedings of the Mystery Swine Disease Committee Meeting, Livestock Conversation Institute, Denver, CO. 1990, 29–31.
- Holtkamp DJ, Polson DD, Torremorrell M. Terminology for classifying swine herds by porcine reproductive and respiratory status. J Swine Health and Prod. 2011, 19:44-56.
- Horter DC, Pogranichniy RM, Chang CC, Evans RB, Yoon KJ, Zimmerman JJ. Characterization of the carrier state in porcine reproductive and respiratory syndrome virus infection. Vet Microbiol. 2002, 86:213-28.
- Loula T. Mystery pig disease. Agri-practice. 1991, 12:23–34.
- Lunney JK, Benfield DA, Rowland RR. Porcine reproductive and respiratory syndrome virus: an update on an emerging and re-emerging viral disease of swine. Virus Res. 2010, 154:1-6.
- Mateu E, Tello M, Coll A, Casal J, Martín M. Comparison of three ELISAs for the diagnosis of porcine reproductive and respiratory syndrome. Vet Rec. 2006, 159:717-8.
- Mateu E, Diaz I. The challenge of PRRS immunology. Vet J. 2008, 177:345-51.
- Meier WA, Galeota J, Osorio FA, Husmann RJ, Schnitzlein WM, Zuckermann FA. Gradual development of the interferon-gamma response of swine to porcine reproductive and respiratory syndrome virus infection or vaccination. Virology. 2003, 309:18-31.
- Nelson EA, Christopher-Hennings J, Benfield DA. Serum immune responses to the proteins of porcine reproductive and respiratory syndrome (PRRS) virus. J Vet Diagn Invest. 1994, 6:410-5.
- Appendices IV and V of the REPORT OF THE OIE AD HOC GROUP ON PORCINE REPRODUCTIVE RESPIRATORY SYNDROME Paris, 9 – 11 June 2008 http://www.oie.int/fileadmin/Home/eng/Our_scientific_expertise/docs/pdf/PRRS_guide_web_bulletin.pdf.
- Rodríguez-Gómez IM, Gómez-Laguna J, Carrasco L. Impact of PRRSV on activation and viability of antigen presenting cells. World J Virol. 2013, 2:146-51.
- Rovira A, Cano JP, Muñoz-Zanzi C. Feasibility of pooled-sample testing for the detection of porcine reproductive and respiratory syndrome virus antibodies on serum samples by ELISA. Vet Microbiol. 2008, 130:60-8.
- Rovira A, Clement T, Christopher-Hennings J, Thompson B, Engle M, Reicks D, Muñoz-Zanzi C. Evaluation of the sensitivity of reverse-transcription polymerase chain reaction to detect porcine reproductive and respiratory syndrome virus on individual and pooled samples from boars. J Vet Diagn Invest. 2007, 19:502-9.
- Rovira A, Reicks D, Muñoz-Zanzi C. Evaluation of surveillance protocols for detecting porcine reproductive and respiratory syndrome virus infection in boar studs by simulation modeling. J Vet Diagn Invest. 2007, 19:492-501.
- Salguero FJ, Frossard JP, Rebel JM, Stadejek T, Morgan SB, Graham SP, Steinbach F. Host-pathogen interactions during porcine reproductive and respiratory syndrome virus 1 infection of piglets. Virus Res. 2015, 202:135-43.
- Segalés J, Domingo M, Balasch M, Solano GI, Pijoan C. Ultrastructural study of porcine alveolar macrophages infected in vitro with porcine reproductive and respiratory syndrome (PRRS) virus, with and without Haemophilus parasuis. J Comp Pathol. 1998, 118:231-43.
- Sur JH, Cooper VL, Galeota JA, Hesse RA, Doster AR, Osorio FA. In vivo detection of porcine reproductive and respiratory syndrome virus RNA by in situ hybridization at different times postinfection. J Clin Microbiol. 1996, 34:2280-6.
- Terpstra C, Wensvoorst G, Pol JMA. Experimental reproduction of porcine epidemic abortion and respiratory syndrome (mystery swine disease) by infection with Lelystad virus: Koch’s postulates fulfilled. Vet Q. 1991, 13:131–36.
- Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu D, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou L, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One. 200, 2:e526.
- Tong GZ, Zhou YJ, Hao XF, Tian ZJ, An TQ, Qiu HJ. Highly pathogenic porcine reproductive and respiratory syndrome, China. Emerg Infect Dis. 2007, 13:1434-6.
Van Alstine WG, Popielarczyk M, Albregts SR. Effect of formalin fixation on the immunohistochemical detection of PRRS virus antigen in experimentally and naturally infected pigs. J Vet Diagn Invest. 2002, 14:504-7. - Vézina SA, Loemba H, Fournier M, Dea S, Archambault D. Antibody production and blastogenic response in pigs experimentally infected with porcine reproductive and respiratory syndrome virus. Can J Vet Res. 1996, 60:94-9.
- Wensvoort G, Terpstra C, Pol JMA, Lask EA, Bloemraad M, de Kluyver EP, Kragten C, van Butten L, den Besten A, Wagenaar F, Broekhuijsen JM, Moonen PJM, Zetstra T, de Boer EA, Tibben AhJ, de Jong MF, van’r Veld P, Groenland GJR, van Gennep JA, Voets MTh, Verheijden JHM, Braamkamp J. Mystery swine disease in the Netherlands: the isolation of Lelystad virus. Vet Q. 1991, 13:121–30.
- Wills RW, Doster AR, Galeota JA, Sur JH, Osorio FA. Duration of infection and proportion of pigs persistently infected with porcine reproductive and respiratory syndrome virus. J Clin Microbiol. 2003, 41:58-62.
- Zimmerman JJ, Benfield DA, Dee SA, Murtaugh MP, Stadejek T, Stevenson GW, Torremorell M. Porcine reproductive and respiratory syndrome virus (porcine arterivirus). In: 10th ed. Diseases of swine, Ed. Wiley-Blackwell. 2012, 31:463-86.