The PRRS virus genetic diversity is the result of mutation and selection, but also recombination:
- Random mutations. The PRRS virus mutation rate is assumed to be the highest so far for a virus, being up to 40 times higher than the mutation rates of well-known viruses, such as Avian influenza virus or Human immunodeficiency virus.
Similarly to other RNA viruses, PRRS virus has an RNA polymerase that does not have the ability to correct the inherent common errors that occur during the transcription of RNA. Since these errors appear every 100-1,000 nucleotides, each new virus may be different from the previous one.
As a consequence, a number of new PRRS viral variants with nucleotide sequences different from the parental one are produced. In infected pigs, PRRS virus exists as a cloud, swarm or distribution of diverse variant viruses related by a similar mutations that collectively contribute to the characteristics of the population and that are subjected to variation, competition and selection.
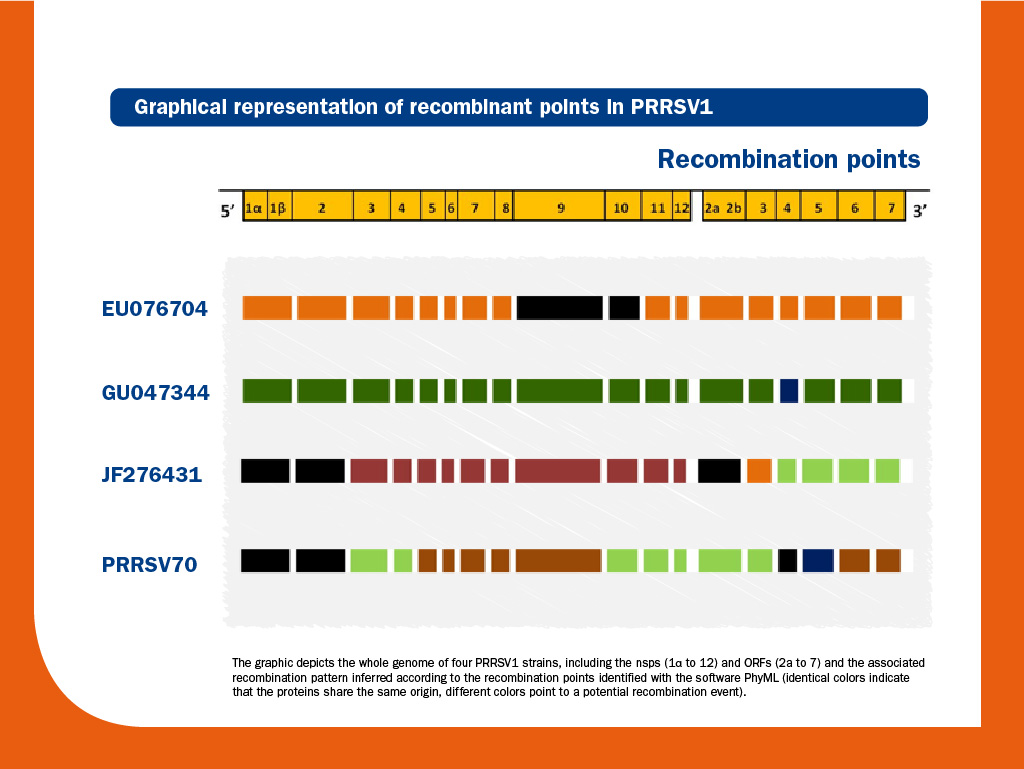
This distribution is known as quasispecies and occurs independently of the strain and the host. Immunomodulation and quasispecies evolution have been speculated to play a crucial role in PRRS virus infection and its chronic/persistent course. - Recombination (exchange of genetic material between two or more strains). In some cases, differences among PRRS virus strains are so great that it is virtually impossible to accept that these differences come from mutation solely.
Similarly, mutations on their own cannot explain the presence of deletions and insertions in numerous locations along the PRRS virus genome. Indeed, when full-length genome sequences have been examined, recombination events have been frequently detected across the genome in both species, pointing this phenomenon as very important to explain PRRS virus genetic diversity.